QIAGEN CLC Genomics
Multi-locus sequence typing (MLST) using QIAGEN CLC Genomics Workbench
1,012 views
• Overview of different tools within QIAGEN CLC Microbial Genomics Module application and research areas supported
• Focused review of isolate typing and characterization Importing data
• Utilization of metadata
• Downloading and managing references
• Database of isolates/ resistances/ MLST
• Walk through of ‘Type a Known Species’ workflow
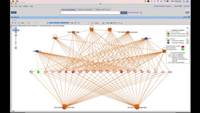
• Review details for each isolate Creating SNP profiles to a specific reference
• Generate a SNP tree for isolate comparison
• Export tabular and high-quality graphical outputs in a wide range of file formats
Related videos
QIAGEN CLC Genomics
QIAGEN CLC Main Workbench training
Day 1: QIAGEN CLC Main Workbench Speaker: Shawn Prince, Sr. Field...
QIAGEN CLC Genomics
Biologically interpret your RNA-seq data with knowledge-based IPA
This webinar will provide an overview of how to analyze data obtained from...
QIAGEN CLC Genomics
QIAGEN CLC Genomics Workbench 20 – scalable desktop software for NGS data analysis
In this webinar, Leif Schauser, Ph.D., Director Product Management Genome...
QIAGEN CLC Genomics
Data analysis on day one - Taxonomic and functional microbiome profiling with the Microbial...
In this webinar, we will introduce the scientist-friendly Microbial Genomics...