QIAGEN CLC Genomics
Metatranscriptome analysis, annotation and pathways investigations using CLC Genomics Workbench
741 views
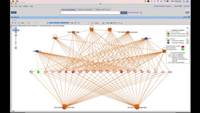
Using CLC Genomics Workbench, we will go through a pipeline for analyzing metatranscriptome NGS data from microbial communities and perform pathways interpretation on it.
· Import "raw" NGS sequencing data and prepare the samples for analysis
· Find relevant annotated genomes with a curated reference database while removing ribosomal RNA with the SILVA database (database of rRNAs)
· De novo assemble the unclassified reads into contigs to also find transcripts of taxa not present in the reference database
· Map the reads to the assembled contigs and database
· Build a functional profile of the samples to get an abundance of GO-terms.
· Statistical analysis of the groups
· Pathway analysis on the differential abundance analysis using MetaCyc database.
· Import "raw" NGS sequencing data and prepare the samples for analysis
· Find relevant annotated genomes with a curated reference database while removing ribosomal RNA with the SILVA database (database of rRNAs)
· De novo assemble the unclassified reads into contigs to also find transcripts of taxa not present in the reference database
· Map the reads to the assembled contigs and database
· Build a functional profile of the samples to get an abundance of GO-terms.
· Statistical analysis of the groups
· Pathway analysis on the differential abundance analysis using MetaCyc database.
Related videos
QIAGEN CLC Genomics
QIAGEN CLC Main Workbench training
Day 1: QIAGEN CLC Main Workbench Speaker: Shawn Prince, Sr. Field...
QIAGEN CLC Genomics
Biologically interpret your RNA-seq data with knowledge-based IPA
This webinar will provide an overview of how to analyze data obtained from...
QIAGEN CLC Genomics
QIAGEN CLC Genomics Workbench 20 – scalable desktop software for NGS data analysis
In this webinar, Leif Schauser, Ph.D., Director Product Management Genome...
QIAGEN CLC Genomics
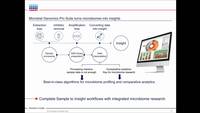
Data analysis on day one - Taxonomic and functional microbiome profiling with the Microbial...
In this webinar, we will introduce the scientist-friendly Microbial Genomics...