Tutorials
Create a SampleSet of Triple-Negative Breast Cancer samples
972 views
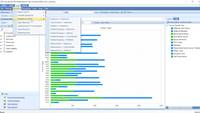
Triple-Negative Breast Cancers can be identified in TCGALand by filtering for clinical variables indicating results of IHC assays. These samples can be sub-grouped within a SampleSet, and this SampleSet grouping can be used to compare samples for differences in expression, mutation frequency, clinical variables, etc.
Interested in learning more?
Click here
Related videos
Tutorials
Identify and Visualize Gene Expression Differences between TNBC and other Breast Tumors
SampleSets can be used to identify "signatures" that reflect differences...
Tutorials
Comparing gene expression with other tumor types
Subtypes of esophageal cancer are derived from different regions of the...
Tutorials
Batch Renaming Files in CLC Workbench
Add/remove characters from element names or replace parts of names,...
Tutorials
Convert Legacy QIAseq Custom Analyses to v24 with QIAseq Panel Analysis Assistance
Custom analyses created with the Analyze QIAseq Samples tool in versions 23.x...